-Search query
-Search result
Showing 1 - 50 of 98 items for (author: roth & bl)
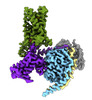
EMDB-43647: 
CryoEM structure of Gi-coupled TAS2R14 with cholesterol and an intracellular tastant
Method: single particle / : Kim Y, Gumpper RH, Roth BL
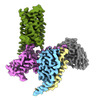
EMDB-43650: 
CryoEM structure of Ggust-coupled TAS2R14 with cholesterol and an intracellular tastant
Method: single particle / : Kim Y, Gumpper RH, Roth BL

EMDB-43656: 
CryoEM structure of Gi-coupled TAS2R14 with cholesterol and an intracellular tastant (Locally refined map)
Method: single particle / : Kim Y, Gumpper RH, Roth BL

EMDB-43657: 
CryoEM structure of Ggust-coupled TAS2R14 with cholesterol and an intracellular tastant (Locally refined map)
Method: single particle / : Kim Y, Gumpper RH, Roth BL

EMDB-35683: 
Dopamine Receptor D1R-Gs-Rotigotine complex
Method: single particle / : Xu P, Huang S, Zhuang Y, Mao C, Zhang Y, Wang Y, Li H, Jiang Y, Xu HE

EMDB-35686: 
Dopamine Receptor D4R-Gi-Rotigotine complex
Method: single particle / : Xu P, Huang S, Zhuang Y, Mao C, Zhang Y, Wang Y, Li H, Jiang Y, Xu HE

EMDB-35684: 
Dopamine Receptor D2R-Gi-Rotigotine complex
Method: single particle / : Xu P, Huang S, Zhuang Y, Mao C, Zhang Y, Wang Y, Li H, Jiang Y, Xu HE

EMDB-35685: 
Dopamine Receptor D3R-Gi-Rotigotine complex
Method: single particle / : Xu P, Huang S, Zhuang Y, Mao C, Zhang Y, Wang Y, Li H, Jiang Y, Xu HE

EMDB-35687: 
Dopamine Receptor D5R-Gs-Rotigotine complex
Method: single particle / : Xu P, Huang S, Zhuang Y, Mao C, Zhang Y, Wang Y, Li H, Jiang Y, Xu HE

EMDB-14986: 
CryoEM structure of Ku heterodimer bound to DNA
Method: single particle / : Hardwick SW, Kefala-Stavridi A, Chirgadze DY, Blundell TL, Chaplin AK

PDB-7zvt: 
CryoEM structure of Ku heterodimer bound to DNA
Method: single particle / : Hardwick SW, Kefala-Stavridi A, Chirgadze DY, Blundell TL, Chaplin AK

EMDB-14955: 
Cryo-EM structure of Ku 70/80 bound to inositol hexakisphosphate
Method: single particle / : Kefala Stavridi A, Chaplin AK, Blundell TL

PDB-7zt6: 
Cryo-EM structure of Ku 70/80 bound to inositol hexakisphosphate
Method: single particle / : Kefala Stavridi A, Chaplin AK, Blundell TL

EMDB-31479: 
Cryo-EM structure of human angiotensin receptor AT1R in complex Gq proteins and Sar1-AngII
Method: single particle / : Zhang D, Xu L
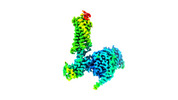
EMDB-29301: 
Neurotensin receptor allosterism revealed in complex with a biased allosteric modulator
Method: single particle / : Krumm BE, Diberto JF, Olsen RHJ, Kang H, Slocum ST, Zhang S, Strachan RT, Fay JF, Roth BL

EMDB-29302: 
CryoEM structure of Go-coupled NTSR1 with a biased allosteric modulator
Method: single particle / : Krumm BE, DiBerto JF, Olsen RHJ, Kang H, Slocum ST, Zhang S, Strachan RT, Fay JF, Roth BL
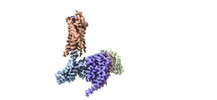
EMDB-29303: 
CryoEM structure of Go-coupled NTSR1
Method: single particle / : Krumm BE, DiBerto JF, Olsen RHJ, Kang H, Slocum ST, Zhang S, Strachan RT, Fay JF, Roth BL
EMDB-26160: 
Structure of human serotonin transporter bound to small molecule '8090 in lipid nanodisc and NaCl
Method: single particle / : Singh I, Seth A

EMDB-33241: 
Cryo-EM Structure of Human Niacin Receptor HCA2-Gi protein complex
Method: single particle / : Yang Y, Kang HJ, Gao RG, Wang JJ, Han GW, DiBerto JF, Wu LJ, Tong JH, Qu L, Wu YR, Pileski R, Li XM, Zhang XC, Zhao SW, Kenakin T, Wang Q, Stevens RC, Peng W, Roth BL, Rao ZH, Liu ZJ

EMDB-29044: 
Structure of Zanidatamab bound to HER2
Method: single particle / : Worrall LJ, Atkinson CE, Sanches M, Dixit S, Strynadka NCJ

PDB-7uih: 
PSMD2 Structure
Method: single particle / : Johnson MC, Bashore C, Ciferri C, Dueber EC

PDB-7ujd: 
PSMD2 Structure bound to MC1 and Fab8/14
Method: single particle / : Johnson MC, Bashore C, Ciferri C, Dueber EC

EMDB-28907: 
Gi bound mu-opioid receptor in complex with beta-endorphin
Method: single particle / : Wang Y, Zhuang Y, DiBerto JF, Zhou XE, Schmitz GP, Yuan Q, Jain MK, Liu W, Melcher K, Jiang Y, Roth BL, Xu HE

EMDB-28908: 
Gi bound mu-opioid receptor in complex with endomorphin
Method: single particle / : Wang Y, Zhuang Y, DiBerto JF, Zhou XE, Schmitz GP, Yuan Q, Jain MK, Liu W, Melcher K, Jiang Y, Roth BL, Xu HE

EMDB-28909: 
Gi bound delta-opioid receptor in complex with deltorphin
Method: single particle / : Wang Y, Zhuang Y, DiBerto JF, Zhou XE, Schmitz GP, Yuan Q, Jain MK, Liu W, Melcher K, Jiang Y, Roth BL, Xu HE

EMDB-28911: 
Gi bound kappa-opioid receptor in complex with dynorphin
Method: single particle / : Wang Y, Zhuang Y, DiBerto JF, Zhou XE, Schmitz GP, Yuan Q, Jain MK, Liu W, Melcher K, Jiang Y, Roth BL, Xu HE

EMDB-28912: 
Gi bound nociceptin receptor in complex with nociceptin peptide
Method: single particle / : Wang Y, Zhuang Y, DiBerto JF, Zhou XE, Schmitz GP, Yuan Q, Jain MK, Liu W, Melcher K, Jiang Y, Roth BL, Xu HE

EMDB-26313: 
C6-guano bound Mu Opioid Receptor-Gi Protein Complex
Method: single particle / : Wang H, Kobilka B

EMDB-25612: 
CryoEM structure of mu-opioid receptor - Gi protein complex bound to mitragynine pseudoindoxyl (MP)
Method: single particle / : Seven AB, Qu Q, Robertson MJ, Wang H, Kobilka BK, Skiniotis G

EMDB-25613: 
CryoEM structure of mu-opioid receptor - Gi protein complex bound to lofentanil (LFT)
Method: single particle / : Seven AB, Qu Q, Huang W, Robertson MJ, Kobilka BK, Skiniotis G

EMDB-27966: 
CryoEM structure of miniGq-coupled hM3Dq in complex with DCZ
Method: single particle / : Zhang S, Fay JF, Roth BL

EMDB-27967: 
CryoEM structure of miniGo-coupled hM4Di in complex with DCZ
Method: single particle / : Zhang S, Fay JF, Roth BL

EMDB-27968: 
CryoEM structure of miniGq-coupled hM3Dq in complex with CNO
Method: single particle / : Zhang S, Fay JF, Roth BL

EMDB-27969: 
CryoEM structure of miniGq-coupled hM3R in complex with Iperoxo
Method: single particle / : Zhang S, Fay JF, Roth BL

EMDB-27970: 
CryoEM structure of miniGq-coupled hM3R in complex with iperoxo (local refinement)
Method: single particle / : Zhang S, Fay JF, Roth BL

EMDB-27752: 
CryoEM structure of Gq-coupled MRGPRX1 with peptide agonist BAM8-22
Method: single particle / : Liu Y, Cao C, Fay JF, Roth BL

EMDB-27753: 
CryoEM structure of Gq-coupled MRGPRX1 with peptide ligand BAM8-22 and positive allosteric modulator ML382
Method: single particle / : Liu Y, Cao C, Fay JF, Roth BL

EMDB-27754: 
CryoEM structure of Gq-coupled MRGPRX1 with ligand Compound-16
Method: single particle / : Liu Y, Cao C, Fay JF, Roth BL

EMDB-25401: 
5-HT2B receptor bound to LSD obtained by cryo-electron microscopy (cryoEM)
Method: single particle / : Barros-Alvarez X, Cao C, Panova O, Roth BL, Skiniotis G

EMDB-25402: 
5-HT2B receptor bound to LSD in complex with heterotrimeric mini-Gq protein obtained by cryo-electron microscopy (cryoEM)
Method: single particle / : Barros-Alvarez X, Kim K, Panova O, Cao C, Roth BL, Skiniotis G

EMDB-25403: 
5-HT2B receptor bound to LSD in complex with beta-arrestin1 obtained by cryo-electron microscopy (cryoEM)
Method: single particle / : Barros-Alvarez X, Cao C, Panova O, Roth BL, Skiniotis G

EMDB-24742: 
PSMD2 with bound macrocycle MC1
Method: single particle / : Johnson MC, Bashore C, Ciferri C, Dueber EC

EMDB-24743: 
PSMD2
Method: single particle / : Johnson MC, Bashore C, Ciferri C, Dueber EC

EMDB-27633: 
Cryo-EM structure of the 5HT2C receptor (INI isoform) bound to lorcaserin
Method: single particle / : Gumpper RH, Fay JF, Roth BL

EMDB-27634: 
Cryo-EM structure of the 5HT2C receptor (INI isoform) bound to psilocin
Method: single particle / : Gumpper RH, Fay JF, Roth BL

EMDB-27635: 
Cryo-EM structure of the 5HT2C receptor (VGV isoform) bound to lorcaserin
Method: single particle / : Gumpper RH, Fay JF, Roth BL

EMDB-27636: 
Cryo-EM structure of the 5HT2C receptor (VSV isoform) bound to lorcaserin
Method: single particle / : Gumpper RH, Fay JF, Roth BL

EMDB-26597: 
CryoEM structure of Go-coupled 5-HT5AR in complex with 5-CT
Method: single particle / : Zhang S, Fay JF, Roth BL

EMDB-26598: 
CryoEM structure of Go-coupled 5-HT5AR in complex with Lisuride
Method: single particle / : Zhang S, Fay JF, Roth BL

EMDB-26599: 
CryoEM structure of Go-coupled 5-HT5AR in complex with Methylergometrine
Method: single particle / : Zhang S, Fay JF, Roth BL
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
